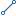| Description | genotype likelihoods comprised of comma separated floating point log10-scaled likelihoods for all possible genotypes given the set of alleles defined in the "Original Sequence Txt" and "Variation Sequence Txt" attributes of "Sequence Variation" entity. In presence of the value in the Genotype attribute the same ploidy is expected and the canonical order is used; without value in Genotype attribute, diploidy is assumed. If A is the allele in "Original Sequence Txt" and B,C,... are the alleles as ordered in "Variation Sequence Txt", the ordering of genotypes for the likelihoods is given by: F(j/k) = (k*(k+1)/2)+j. In other words, for biallelic sites the ordering is: AA,AB,BB; for triallelic sites the ordering is: AA,AB,BB,AC,BC,CC, etc.
For example: GT:GL 0/1:-323.03,-99.29,-802.53 |
 Assay / Sequence Variation Sk
Assay / Sequence Variation Sk Effective From Dt
Effective From Dt Effective To Dt
Effective To Dt Expected Alternate Allele Counts Txt
Expected Alternate Allele Counts Txt Genotype Filter
Genotype Filter Genotype Likelihoods of Heterogeneous Ploidy Txt
Genotype Likelihoods of Heterogeneous Ploidy Txt Genotype Likelihoods Txt
Genotype Likelihoods Txt Genotype Quality Txt
Genotype Quality Txt Genotype Txt
Genotype Txt Haplotype Qualities Txt
Haplotype Qualities Txt Load Info Sk
Load Info Sk Mapping Quality Num
Mapping Quality Num Phase Set Txt
Phase Set Txt Phasing Quality Txt
Phasing Quality Txt Phred-Scaled Genotype Likelihoods Txt
Phred-Scaled Genotype Likelihoods Txt Phred-Scaled Genotype Posterior Probabilities Txt
Phred-Scaled Genotype Posterior Probabilities Txt Read Depth Num
Read Depth Num Source Code Sk
Source Code Sk Tenant Sk
Tenant Sk Valid From Ts
Valid From Ts Valid To Ts
Valid To Ts